Displacement Loop (D-loop) detail |
 |
Mitochondrial DNA Map D-LoopBiochemical Society Transaction (2000) 28, 154-159 |
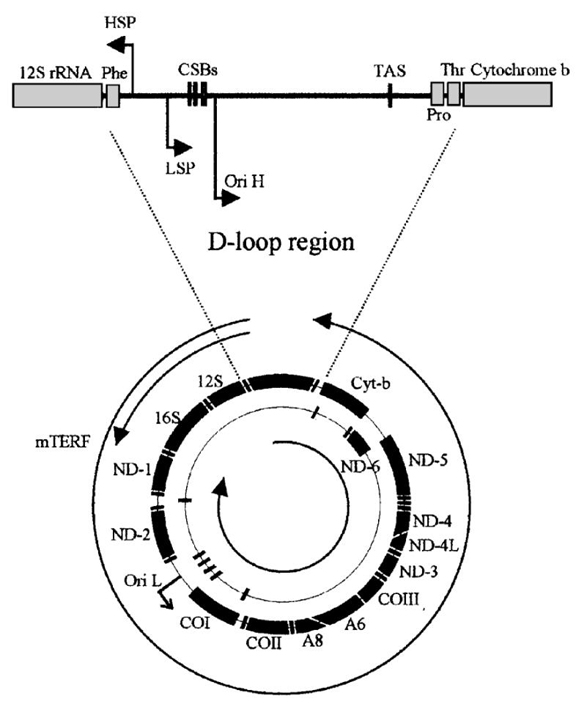
Figure 1 Schematic diagram of mtDNA, the major polycistronic RNAs and the displacement (D-) loop region and its cis-acting elements
The human mitochondrial genome is 16569 bp. The bold outer circle is representative of the heavy (H-) strand and the bold inner circle the light (L-) strand. Ori H and Ori L denote origins of replication and arrows indicate direction. Transcript-initiation sites are denoted HSP and LSP and their direction of elongation indicated by arrows. The three main polycistronic RNA species are shown as the two outer and one inner thin black lines. The premature termination site of HSP-initiated transcripts is denoted by mTERF at the 3'-end of the 16 S rRNA gene. Peptide and rRNA genes are denoted by bold black lines and annotation and the tRNA genes are denoted by bold thick black lines but are not annotated. The D-loop region is shown in closer detail. The conserved sequence blocks (CSBs), are the sites of LSP-initiated transcript maturation for DNA replication. Small RNA species initiated from LSP are used to prime DNA synthesis. Nascent DNA species are often terminated downstream of the termination-associated sequence (TAS) site. This premature replication termination produces the 7 S DNA D-loop. Subunit abbreviations are: ND, NADH dehydrogenase; CO, cytochrome c oxidase; A, ATP synthetase; Cyt-b, apocytochrome b.